JBioFramework: Simulation for Education
Overview
Welcome to JBioFramework!
JBioFramework (JBF) is a collection of chemical separations simulations (Electrophoresis, Chromatography and Mass Spectrometry) that can enable students to experience the interface and results with these techniques in a safe, free environment. A full collection of the simulations can be downloaded from Sourceforge. It is written in the Java programming language and will run on computers with operating systems that have the Java Virtual Machine installed.
What's New?
We have a new release of the 1D electrophoresis simulation that has been expanded from 2 wells to 10 wells. The user can now load their own protein mixtures for separation, using the FASTA format for protein sequence. It is possible to simulate the results of a protein purification process, even with thicker bandwidths for proteins as they are purified. Here is an example of a screenshot. Clicking on the image will bring up the fully functioning application.
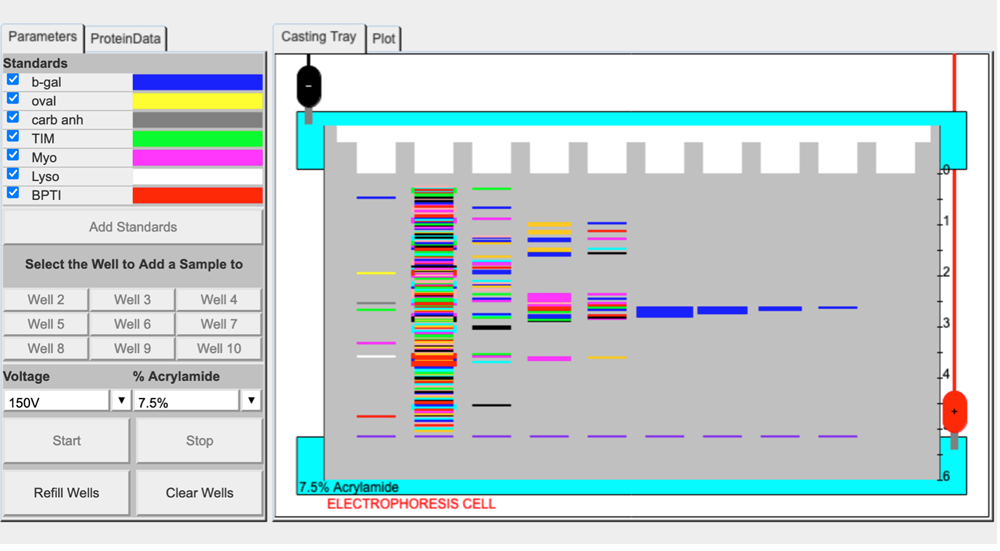
This simulation of 1D electrophoresis can be directly linked to protein datasets for the BASIL curriculum, but it can also be linked to custom designed protein datasets, problem sets and in silico experiments to be used for classroom instruction, virtual labs or pre-lab preparation. If you are interested in learning how to design your own datasets or would like to collaborate in some other fashion, email Paul Craig.
Presentations
Bernhardt, T., Seyfarth, M., Hanson, R.M., Roberts, R. and Craig, P.A. (2022), An Enhanced 1D Electrophoresis Simulation with Pedagogical Tools. The FASEB Journal, 36:. https://doi.org/10.1096/fasebj.2022.36.S1.0R332, ASBMB Annual Meeting, Philadelphia, PA, April 2022.
Emily R. Sekera, Aidan Sawyer, Bader Alharbi, Paul A. Craig. A Wet Lab Simulation to Enhance the Learning Experience of Undergraduates, ACS Regional Meeting at RIT, April 2014.
Emily R. Sekera, Aidan Sawyer, Paul A. Craig. A 2DE-Tandem MS Simulation with a Structural Interface, ASBMB Annual Meeting in Boston, MA, April 2013.
Amanda Fisher, Paul Craig, Emily Sekera. A Mass Spectrometry Simulation for Biochemistry Education, ASBMB Annual Meeting in San Diego, CA, April 2012.
Amanda Fisher, Kyle Dewey, Paul Craig. Developing and Sustaining Scientific Software Applications for Biochemistry Education, ASBMB Annual Meeting in Washington, DC, April 2011.